Kidney Studies

A Novel, Translatable Platform for Renal Research
Pebble's LIVING-KIDNEY System
The Pebble LIVING-KIDNEY system replicates in-vivo physiology, circulating warm, nutrient rich, oxygenated blood through arteries and deoxygenated, nutrient depleted blood through veins. This restores metabolic function within the kidney, providing an ideal platform for testing renal therapies in a translatable model.
Perfusion Protocols
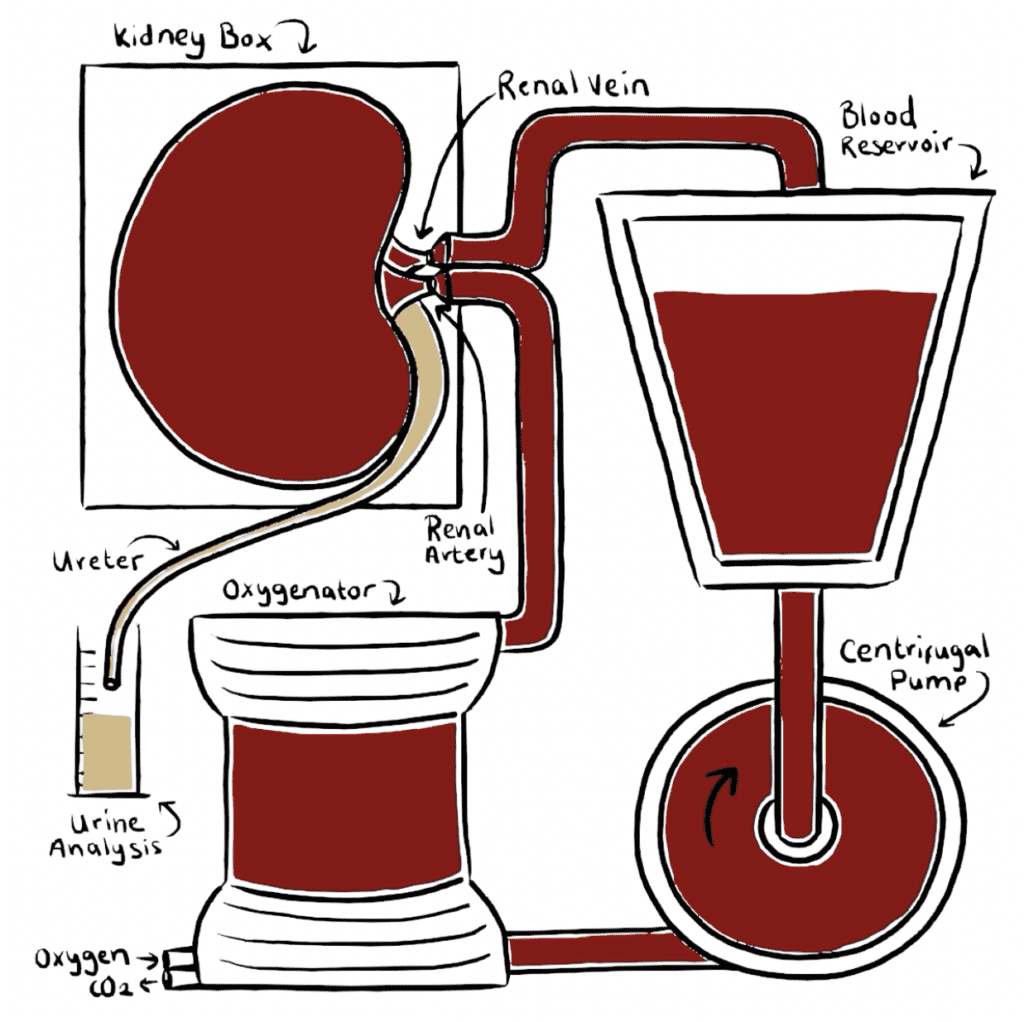
Kidneys are perfused with autologous blood and maintained under physiological conditions.
Our LIVING-KIDNEY system reproduces the living environment, providing an excellent platform for translatable testing. A warm, oxygenated blood-based perfusate is pumped through the renal vasculature, providing nutrients and restoring full function & metabolism. This means kidneys filter the blood and produce urine as they would inside the body.
We only use clinical gold standard protocols for organ preservation (including approved cold-storage and machine perfusion approaches). All solutions, consumables and hardware are FDA/MHRA/EMA approved.
Our expert multidisciplinary team are highly experienced in the field of kidney perfusion, so if you would like to test your innovation, please get in touch.
Renal Evaluation
LIVING-KIDNEY System Models
- Simulating acute renal failure scenarios to evaluate potential therapies.
- Monitoring biomarker changes in response to induced injury and subsequent recovery.
- Assessing the effectiveness of renoprotective agents in acute injury settings.
- Studying the pharmacokinetics of drugs, including metabolism and clearance rates.
- Investigating the impact of renal impairment on drug metabolism.
- Testing the renal excretion of novel pharmacological compounds.
- Evaluating the effect of antihypertensive drugs on renal function and blood pressure regulation.
- Analysing the role of kidneys in the pathophysiology of hypertension.
- Assessing drug-induced changes in the renin-angiotensin-aldosterone system
- Replicating inflammatory conditions of the glomeruli to assess therapeutic approaches.
- Studying the immune response within the kidney during glomerular inflammation.
- Assessing the efficacy of anti-inflammatory and immunosuppressive treatments.
- Testing the renal toxicity of drugs, chemicals, and environmental toxins.
- Assessing protective strategies against known nephrotoxic substances.
- Using biomarkers to detect early signs of renal damage.
- Simulating the effects of reduced blood flow followed by restoration to study potential damage and repair mechanisms.
- Evaluating therapeutic agents designed to mitigate ischemia-reperfusion injuries.
- Testing the efficacy of immunomodulatory therapies in reducing transplant rejection.
- Monitoring the biomarkers associated with graft health and rejection.
- Assessing new surgical techniques and preservation solutions for renal transplantation.
Therapeutic Evaluation
- Evaluating the efficacy of various drugs on renal function, including diuretics, anti-hypertensives, and antibiotics.
- Assessing dose-response relationships for different pharmacological agents to optimise therapeutic dosing.
- Monitoring side effects and systemic impacts of pharmacological treatments on kidney health.
- Conducting time-course studies to understand the onset, duration, and peak of drug action.
- Investigating the mechanisms of action of pharmacological agents through cellular and molecular studies.
- Testing the effectiveness of biologic agents like monoclonal antibodies or recombinant proteins on renal diseases.
- Assessing the kidney’s response to cytokines or growth factors used in biological therapies.
- Monitoring the renal effects of gene therapies, including vector distribution and expression.
- Studying the long-term renal outcomes and regenerative potential of biological therapies.
- Investigating the potential for stem cell therapies to repair or regenerate renal tissue.
- Assessing the ability of tissue-engineered constructs to integrate with and restore kidney function.
- Evaluating the biocompatibility and acute survival of implanted cells or tissues.
- Developing and testing biomaterials for their effectiveness in supporting renal tissue regeneration.
- Assessing the efficacy of immunosuppressants in preventing renal transplant rejection or treating autoimmune renal diseases.
- Investigating the kidney’s response to immunostimulatory agents in immunocompromised states.
- Evaluating the renal effects of modulation of inflammatory pathways using cytokines or small molecule inhibitors.
- Testing the ability of immunotherapies to target renal cancer cells without harming healthy tissue.
- Monitoring the impact of immunomodulatory drugs on renal immune cell populations and function.
- Evaluating the selectivity and efficacy of small molecule inhibitors on renal cancer or disease pathways.
- Testing the effects of targeted gene silencing techniques like siRNA on renal disease models.
- Assessing the delivery and efficacy of targeted nanoparticle systems in renal tissues.
- Investigating the impact of hormone therapies on renal diseases, such as those targeting the Renal-Angiotensin-Aldosterone-System.
- Analysing the renal outcomes of precision targeting based on genetic or biomarker profiling.
- Assessing the effectiveness of antioxidants and other protective agents against renal oxidative stress.
- Evaluating the role of dietary and lifestyle interventions in maintaining renal health and preventing disease.
- Testing the protective effects of novel compounds against nephrotoxins and other renal insults.
- Investigating the renal protective benefits of blood pressure control strategies.
- Analysing the potential for renal protective effects of modifying lipid metabolism.
- Tailoring pharmacological and biological therapies based on individual genetic and biomarker profiles.
- Assessing the impact of patient-specific factors on the efficacy and toxicity of renal therapies.
- Developing predictive models to forecast individual responses to renal treatments.
- Evaluating the use of patient-derived organoids or tissues for personalised therapeutic screening.
- Implementing advanced diagnostics, including genomic and proteomic analyses, to guide personalised renal care.
- Screening potential nephrotoxic effects of new drugs using in vitro and ex vivo kidney models.
- Evaluating the protective effects of co-therapies intended to mitigate drug-induced renal damage.
- Conducting long-term toxicity studies to assess the chronic impact of drugs on renal function.
- Monitoring biomarkers of renal injury during drug exposure to assess toxicity risk.
- Developing high-throughput assays to rapidly screen a wide range of compounds for renal toxicity.

Functional Analysis
- Filtration Efficiency: Assaying glomerular selectivity for size and charge to understand filtration barriers.
- Concentration Capacity: Gauging the kidney’s urine-concentrating ability for fluid balance and osmoregulation.
- Clearance Rate: Quantitative analysis of renal clearance for a spectrum of substances to inform kidney excretory performance.
- Plasma Flow: Measuring renal perfusion metrics for insights into kidney health and function.
- Tubular Reabsorption: Investigating reabsorption dynamics for critical solutes and water, mirroring tubular health.
- Secretory Functions: Evaluating secretion of ions and waste products, vital for acid-base and electrolyte balance.
- Threshold Determination: Defining plasma concentration thresholds for various solutes before renal excretion occurs.
- Pressure Natriuresis/Diuresis: Probing sodium and water excretion responses to blood pressure changes.
- Aldosterone Response: Assessing kidney’s reaction to aldosterone for sodium-potassium balance.
- Functional Reserve: Testing the kidney’s ability to augment function under stress or disease.
- Artery Flow Measurement: Quantifying blood entry rates into the renal artery, indicative of perfusion status.
- Pressure Monitoring: Continuous arterial pressure tracking to understand kidney stress responses.
- Intra-Renal Resistance: Evaluating intrarenal vascular resistance, indicative of health or disease states.
- Vascular Compliance: Assessing renal blood vessel elasticity for accommodating flow alterations.
- Flow-Pressure Response: Investigating autoregulation via renal blood flow and pressure changes.
- Segmental Arterial Analysis: Segment-specific flow pattern differentiation for regional perfusion insights.
- Resistance Index Mapping: Charting resistance across renal vascular territories for pathophysiological insights.
- IR Thermal Imaging: Utilising regional thermal analysis for perfusion heterogeneity insights.
- Creatinine Monitoring: Tracking serum and urine creatinine for renal function status.
- Albumin Quantification: Measuring urine albumin as a sensitive glomerular health indicator.
- NGAL: Detecting early kidney injury markers NGAL in plasma and urine.
- KIM-1: Quantifying tubular injury marker KIM-1 levels.
- Cystatin C Analysis: Employing Cystatin C as a GFR and kidney dysfunction biomarker.
- Beta-2 Microglobulin: Beta-2 Microglobulin as a tubular injury indicator.
- NAG Activity: Assaying urinary NAG for tubular cell damage.
- TFF3: Investigating TFF3 as a marker for tubular injury and repair.
- FeNa: Assessing sodium filtration and reabsorption efficiency via FeNa.
- Protein Profiling: Monitoring abnormal urinary protein excretion patterns.
- Albumin Creatinine Ratio (ACR) Tracking: Utilising ACR for early renal effects of therapies.
- Drug/Metabolite Excretion: Pharmacokinetic profiling of therapeutic agent clearance in urine.
- Osmolality Response: Assessing treatments on urine concentration capabilities.
- Proteomic/Metabolomic Signatures: Profiling urine for specific changes related to therapy action or device interaction.
- Gene Therapy Biomarkers: Identifying gene therapy-specific urinary biomarkers.
- Nephrotoxicity Evaluation: Quantifying renal toxicity markers after exposure to potential nephrotoxins.
- pH Variation Analysis: Tracking urine pH shifts resulting from metabolic therapy effects.
- Electrolyte Disruption: Evaluating drug or cell therapy impacts on renal electrolyte regulation.
- Cell Therapy Metabolic Assessment: Monitoring urinary metabolic by-products from cellular therapies.
- Device Impact Cytology: Analysing urine sediment for medical device effects on renal cells.
- Therapeutic Agent Stability: Testing biologic drug stability through renal processing.
- Vector Shedding: Monitoring gene therapy vector clearance via urine as a biodistribution measure.
- Nanotherapeutic Filtration: Assessing renal filtration of nanoparticles and nanoformulated drugs.
- Immune Activation Markers: Detecting immunomodulatory drug effects through urinary markers.
- Device Erosion Sediment Analysis: Searching for device erosion signs in urine sediment.
- ATMP Functional Biomarkers: Leveraging urine biomarkers for ATMP efficacy and safety.
- Urinary Exosomes: Analysing urinary exosomes for insights into cell therapy outcomes.
- Pharmacodynamics: Studying kidney responses to targeted renal therapies.
- Sodium Balance: Measuring serum and urine sodium for volume and osmotic regulation insights.
- Potassium Regulation: Monitoring potassium management by the kidney in various states.
- Calcium/Phosphorus Homeostasis: Assessing renal calcium and phosphorus management, including vitamin D’s role.
- Chloride Concentration: Clarifying renal chloride management for acid-base balance insights.
- Bicarbonate Reabsorption: Investigating renal bicarbonate management and its role in pH regulation.
- Anion Gap Assessment: Utilizing anion gap measures for metabolic disturbance insights.
- Acid Load Handling: Testing renal responses to acid challenges for metabolic function insights.
- Diuretic Response: Studying diuretic impacts on renal electrolyte balance.
- Trace Element Handling: Evaluating renal regulation of trace elements and its implications.
- Blood Gas Analysis: Profiling blood pH, CO2, and bicarbonate for renal compensatory capacity insights.
- Base Excess/Deficit: Measuring base excess or deficit for renal acid-base equilibrium contributions.
- Urine pH Measurement: Assessing urine pH for renal acidification or alkalization capacity.
- Ammoniagenesis Evaluation: Quantifying renal ammonium production for systemic pH balance contributions.
- RTA Analysis: Differentiating renal tubular acidosis types and their underlying mechanisms.
- Buffer System Function: Probing renal buffer systems for plasma pH maintenance.
- Bicarbonate Handling Assessment: Measuring bicarbonate reabsorption for renal metabolic acidosis correction insights.
- Respiratory Compensation Profiling: Charting renal responses to respiratory acidosis or alkalosis.
- Acidification Test: Conducting acidification tests for renal acid handling capacity.
- Acid Adaptation Monitoring: Tracking renal adaptive responses to increased acid loads.
- Tissue pO2 Measurements: Monitoring intrarenal oxygen levels for hypoxia response insights.
- Hypoxia Inducible Factor (HIF) Pathway Analysis: Probing HIF pathways for erythropoietin production and hypoxia response insights.
- OCR in Renal Tissue: Assessing metabolic demand through renal oxygen consumption rates.
- Oxygen Gradient Mapping: Establishing intrarenal oxygen gradients for understanding renal hypoxia adaptations.
- Filtration Rate Assessment: Evaluating GFR variations for renal adaptability insights.
- Hemodynamic Influence: Profiling blood pressure impacts on GFR for autoregulation insights.
- Filtration Marker Comparison: Employing various markers to assess GFR accuracy.
- Clearance Studies: Conducting GFR measurements using renal clearance of exogenous substances in research settings.
- Electrolyte Reabsorption: Assessing tubular reabsorption efficiency for key electrolytes.
- Glucose Handling: Measuring tubular glucose reabsorption for diabetic kidney disease insights.
- Acid-Base Regulation: Evaluating tubular secretion of hydrogen ions and bicarbonate reabsorption.
- Waste Product Secretion: Examining tubular waste and foreign substance secretion for drug clearance insights.
- Concentration/Dilution Capacity: Testing renal tubular urine concentration and dilution in response to hydration status.
- Leukocyte State: Analysing the activation status and subtypes of leukocytes present in the kidney and circulating perfusate to determine immune readiness and potential inflammatory responses.
- Blood-Perfusate Cytokine Concentrations: Measuring the levels of various cytokines in the perfusate to assess the inflammatory state and identify specific immune signalling pathways being activated.
- Complement Activation: Assessing the activation of the complement system as a part of innate immunity and its role in inflammation and renal injury.
- Antibody Response: Evaluating the generation of specific antibodies as a marker of immune response to infections, autoimmunity, or therapeutic interventions.
- Renal Leukocyte Extravasation: Monitoring the movement of white blood cells from the bloodstream into renal tissue, an essential process for immune surveillance and response to injury or infection.
- T Cell Profiling: Characterising T cell populations, including helper, cytotoxic, regulatory, and memory subsets, to understand adaptive immune responses in renal contexts.
- Macrophage Activation Status: Quantifying the activation state and polarization (M1 vs. M2) of macrophages within kidney tissues, which can influence inflammation and fibrosis.
- Dendritic Cell Maturation: Assessing the maturation and antigen-presenting capacity of dendritic cells, which are key for initiating T cell responses.
- Natural Killer (NK) Cell Activity: Evaluating the cytotoxic activity of NK cells towards infected or transformed cells within the renal environment.
- B Cell Maturation and Antibody Production: Assessing B cell maturation and the capacity for specific antibody production, critical for humoral immunity.
- Immune Checkpoint Expression: Analysing the expression of immune checkpoint molecules like PD-1 and CTLA-4 which can modulate the amplitude of immune responses.
- Inflammatory Mediator Profiling: Quantifying soluble mediators such as chemokines, acute phase reactants, and adhesion molecules that facilitate immune cell migration and activation.
- Autoantibody Identification: Detecting the presence of autoantibodies that can signal autoimmune conditions affecting the kidneys.
- Cytokine Storm Assessment: Evaluating the propensity for a cytokine storm, a potentially fatal immune response that can result from therapeutic interventions or infections.
- Pathogen-Specific Immune Response: Characterizing the immune response to specific pathogens that can affect kidney health, including bacteria, viruses, and fungi.
- Immune Memory Assessment: Investigating the presence of memory immune cells to assess the long-term immunity and potential for quick response to previously encountered antigens.
Structural Assessment
- Assessing organ size, shape, and colour for abnormalities or pathologies.
- Evaluating surface characteristics such as texture and the presence of lesions or growths.
- Measuring organ weight to detect changes indicative of disease or treatment effects.
- Conducting palpation to determine consistency and detect areas of fibrosis or infarction.
- Using perfusion imaging techniques to assess vascular architecture and blood flow.
- Preparing tissue sections for examination under a microscope to observe cellular structure and organization.
- Staining tissues with haematoxylin and eosin (H&E) to highlight general tissue architecture.
- Employing special stains like Masson’s trichrome for fibrosis or periodic acid-Schiff for basement membranes.
- Quantifying cellular changes such as hypertrophy, hyperplasia, or atrophy.
- Detecting and characterizing pathological changes like inflammation, necrosis, or neoplasia.
- Determining rates of oxygen consumption and carbon dioxide production for metabolic rate analysis.
- Measuring ATP levels and mitochondrial function to assess cellular energy status.
- Quantifying metabolite levels in tissue extracts using techniques like NMR or mass spectrometry.
- Assaying enzyme activities involved in key metabolic pathways such as glycolysis or Krebs cycle.
- Investigating the effects of metabolic modulators on tissue substrate utilization.
- Using specific antibodies to visualize the distribution of proteins or antigens within tissues.
- Quantifying the expression levels of markers for cell proliferation, such as Ki-67.
- Detecting the presence and localization of cytokines, growth factors, or receptors.
- Visualizing the distribution of immune cells within tissues to assess inflammatory responses.
- Employing fluorescence or confocal microscopy to obtain high-resolution images of labelled antigens.
- Conducting ultrasound imaging for non-invasive assessment of organ structure and blood flow.
- Using optical coherence tomography for high-resolution imaging of tissue microstructures.
- Implementing two-photon microscopy to image live tissues at cellular and subcellular levels.
- Measuring tissue elasticity and stiffness using techniques such as elastography.
- Conducting tensile and compressive tests to evaluate mechanical properties of tissues.
- Using indentation testing to assess the hardness and resilience of tissues.
- Performing shear tests to understand the viscoelastic properties of organ tissue.
- Assessing the impact of diseases or treatments on the biomechanical integrity of tissues.
- Analysing tissue extracts for specific biochemical markers like electrolytes or lipids.
- Measuring the activity of enzymes relevant to tissue function or pathology.
- Assessing the levels of metabolites that are indicative of organ health or disease states.
- Conducting assays for oxidative stress markers such as malondialdehyde or glutathione.
- Quantifying the presence of small molecules or nutrients within the tissue.
- Evaluating endothelial function and the presence of atherosclerotic changes within vessels.
- Measuring vascular reactivity to vasoactive substances to assess endothelial health.
- Assessing angiogenic response in tissue samples in the context of growth or repair.
- Quantifying capillary density and morphology to evaluate tissue perfusion and oxygenation.
- Investigating the role of the vasculature in organ-specific diseases and their response to treatments.
Molecular Biology
- This involves sequencing the entire genome of the kidney tissue to identify variations in DNA sequence, such as single nucleotide polymorphisms (SNPs), copy number variations (CNVs), and structural rearrangements.
- Genomic analysis can provide insights into genetic predispositions to kidney diseases, as well as the molecular mechanisms underlying renal physiology and pathology.
- Transcriptomic analysis involves quantifying and profiling the entire transcriptome of the kidney tissue, including messenger RNA (mRNA), non-coding RNA (such as microRNA and long non-coding RNA), and splice variants.
- This can help identify differentially expressed genes, alternative splicing events, and regulatory networks involved in kidney development, function, and disease.
- Epigenomic analysis examines the epigenetic modifications (e.g., DNA methylation, histone modifications, chromatin accessibility) that regulate gene expression patterns in the kidney tissue.
- Understanding epigenetic mechanisms can provide insights into how environmental factors influence renal health and disease susceptibility.
- Proteomic analysis aims to characterize the entire complement of proteins expressed in the kidney tissue, including post-translational modifications (PTMs) and protein-protein interactions.
- This can reveal changes in protein expression levels, modifications, and interactions associated with renal diseases, drug responses, and physiological processes.
- Metabolomic analysis involves profiling the small molecule metabolites present in the kidney tissue, providing insights into metabolic pathways, cellular processes, and metabolic signatures associated with renal health and disease states.
- Metabolomics can help identify biomarkers of kidney dysfunction, as well as metabolic alterations induced by drug treatments or environmental exposures.
- Lipidomic analysis focuses on the comprehensive profiling of lipids in the kidney tissue, including lipid species, classes, and lipid metabolism pathways.
- This can uncover lipid biomarkers of kidney diseases, as well as lipid-mediated signaling pathways involved in renal homeostasis and pathogenesis.
- Glycomic analysis characterises the entire repertoire of glycans and glycoproteins in the kidney tissue, revealing glycan structures, modifications, and glycosylation patterns.
- This can provide insights into glycan-mediated interactions, cell signaling, and glycosylation changes associated with renal diseases and physiological processes.
- Profile gene expression patterns within the kidney tissue using techniques such as RNA sequencing or quantitative PCR to identify changes associated with disease or experimental interventions.
Pebble's Customisable
Service Package
Why Choose Pebble?
Testimonials

Prof Marc Clancy
Consultant Transplant Surgeon, Greater Glasgow and Clyde NHS Trust
“Our team used Pebble when we needed to get a clear picture of where our clinical kidney preservation protocol might be falling short. Whilst adequate for viability assessment, our established EVNP protocols had proved unsuitable for the longer term preservation we need to get the best results from modern day donor organs.
Their team got straight to work, comparing different approaches in a practical study, then crunching the numbers to show us what was going on. They delivered a report that identified several limitations to the clinical protocol, pointing out where we could make improvements.
Thanks to Pebble, we are making real changes to how we handle donor kidneys and hope to improve their benefit in our patients. I’m confident this will lead to real clinical improvements for these patients in the future. It’s been a solid partnership that’s making a real difference in our work, and we expect this to continue to grow.”

